scipy.optimize.minimize(fun, x0, args=(), method='L-BFGS-B', jac=None, bounds=None, tol=None, callback=None, options={'disp': None, 'maxls': 20, 'iprint': -1, 'gtol': 1e-05, 'eps': 1e-08, 'maxiter': 15000, 'ftol': 2.220446049250313e-09, 'maxcor': 10, 'maxfun': 15000})```
The argument `fun` is a callable function that returns the misfit between the simulated and the observed data. If `jac` is a Boolean and is `True`, `fun` is assumed to return the gradient along with the objective function - as is our case when applying the adjoint-state method.
## Dask
[Dask](https://dask.pydata.org/en/latest/#dask) is task-based parallelization framework for Python. It allows us to distribute our work among a collection of workers controlled by a central scheduler. Dask is [well-documented](https://docs.dask.org/en/latest/), flexible, an currently under active development.
In the same way as in [04_dask.ipynb](https://github.com/devitocodes/devito/blob/main/examples/seismic/tutorials/04_dask.ipynb), we are going to use it here to parallelise the computation of the functional and gradient as this is the vast bulk of the computational expense of FWI and it is trivially parallel over data shots.
## Forward modeling
We define the functions used for the forward modeling, as well as the other functions used in constructing and deconstructing Python/Devito objects to/from binary data as follows:
::: {#cell-8 .cell execution_count=1}
``` {.python .cell-code}
# NBVAL_IGNORE_OUTPUT
import numpy as np
from scipy import optimize
from distributed import Client, LocalCluster, wait
import cloudpickle as pickle
# Import acoustic solver, source and receiver modules.
from examples.seismic import demo_model, AcquisitionGeometry, Receiver
from examples.seismic.acoustic import AcousticWaveSolver
# Import convenience function for plotting results
from examples.seismic import plot_image
# Set up inversion parameters.
param = {
't0': 0.,
'tn': 1000., # Simulation last 1 second (1000 ms)
'f0': 0.010, # Source peak frequency is 10Hz (0.010 kHz)
'nshots': 5, # Number of shots to create gradient from
'shape': (101, 101), # Number of grid points (nx, nz).
'spacing': (10., 10.), # Grid spacing in m. The domain size is now 1km by 1km.
'origin': (0, 0), # Need origin to define relative source and receiver locations.
'nbl': 40 # nbl thickness.
}
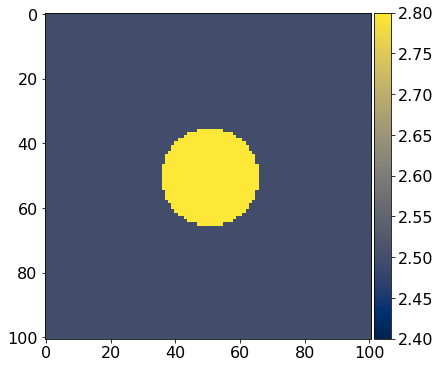
def get_true_model():
''' Define the test phantom; in this case we are using
a simple circle so we can easily see what is going on.
'''
return demo_model('circle-isotropic', vp_circle=3.0, vp_background=2.5,
origin=param['origin'], shape=param['shape'],
spacing=param['spacing'], nbl=param['nbl'])
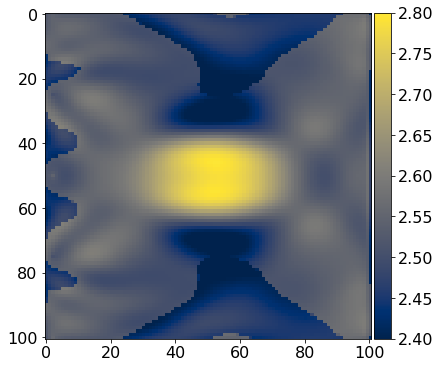
def get_initial_model():
'''The initial guess for the subsurface model.
'''
# Make sure both model are on the same grid
grid = get_true_model().grid
return demo_model('circle-isotropic', vp_circle=2.5, vp_background=2.5,
origin=param['origin'], shape=param['shape'],
spacing=param['spacing'], nbl=param['nbl'],
grid=grid)
def wrap_model(x, astype=None):
'''Wrap a flat array as a subsurface model.
'''
model = get_initial_model()
v_curr = 1.0/np.sqrt(x.reshape(model.shape))
if astype:
model.update('vp', v_curr.astype(astype).reshape(model.shape))
else:
model.update('vp', v_curr.reshape(model.shape))
return model
def load_model(filename):
""" Returns the current model. This is used by the
worker to get the current model.
"""
with open(filename, 'rb') as fh:
pkl = pickle.load(fh)
return pkl['model']
def dump_model(filename, model):
''' Dump model to disk.
'''
with open(filename, "wb") as fh:
pickle.dump({'model': model}, fh)
def load_shot_data(shot_id, dt):
''' Load shot data from disk, resampling to the model time step.
'''
with open(f"shot_{shot_id}.p", "rb") as fh:
pkl = pickle.load(fh)
return pkl['geometry'], pkl['rec'].resample(dt)
def dump_shot_data(shot_id, rec, geometry):
''' Dump shot data to disk.
'''
with open(f'shot_{shot_id}.p', "wb") as fh:
pickle.dump({'rec': rec, 'geometry': geometry}, fh)
def generate_shotdata_i(param):
""" Inversion crime alert! Here the worker is creating the
'observed' data using the real model. For a real case
the worker would be reading seismic data from disk.
"""
# Reconstruct objects
with open("arguments.pkl", "rb") as cp_file:
cp = pickle.load(cp_file)
solver = cp['solver']
# source position changes according to the index
shot_id = param['shot_id']
solver.geometry.src_positions[0, :] = [20, shot_id*1000./(param['nshots']-1)]
true_d = solver.forward()[0]
dump_shot_data(shot_id, true_d.resample(4.0), solver.geometry.src_positions)
def generate_shotdata(solver):
# Pick devito objects (save on disk)
cp = {'solver': solver}
with open("arguments.pkl", "wb") as cp_file:
pickle.dump(cp, cp_file)
work = [dict(param) for i in range(param['nshots'])]
# synthetic data is generated here twice: serial(loop below) and parallel (via dask map functionality)
for i in range(param['nshots']):
work[i]['shot_id'] = i
generate_shotdata_i(work[i])
# Map worklist to cluster, We pass our function and the dictionary to the map() function of the client
# This returns a list of futures that represents each task
futures = c.map(generate_shotdata_i, work)
# Wait for all futures
wait(futures)